Surfaceome Profiling of Rhabdomyosarcoma Reveals Target Antigens for Immune-Based Therapies
Roxane R. Lavoie, R.A., Fabrice Lucien, PhD, Vidhu Joshi, MD, Patricio C. Gargollo, MD, Candace Granberg, M.D..
Mayo Clinic, Rochester, MN, USA.
BACKGROUND: Rhabdomyosarcoma (RMS) is the most common soft tissue tumor in children, with nearly 20% of children presenting with locally aggressive and/or metastatic disease. Despite protocol-based chemotherapy and radiation, the 5-year overall survival rate is still less than 40%. Additionally, the late and long-term side effects of treatment in survivors of RMS affect multiple organs and systems, and can take years to develop. Among the two main RMS subtypes, alveolar (ARMS) and embryonal (ERMS), ARMS is associated with a poorer outcome. This is mainly justified by abnormal expression of the PAX3/7-FOXO1 fusion gene (PF) leading to aberrant transcriptional activation of genes involved in cell proliferation, invasion and metastasis formation. Targeting PAX3/7-FOXO1 downstream effectors represents a promising avenue for the development of immune-based therapies including neutralizing antibodies, oncolytic viruses or CAR T-therapy.
METHODS: Our group aims to identify tumor-specific antigens expressed at the cell-surface of RMS tumor cells. Aberrant expression of cell-surface proteins may represent an important source of anti-cancer targets since their extracellular accessibility makes therapeutic intervention more effective with immunotherapy. By using cell-surface isolation and targeted proteomic profiling by mass spectrometry, we have analyzed the "surfaceome" of fusion-positive (FP), fusion-negative (FN) patient-derived RMS cells and normal skeletal muscle. We performed high-throughput protein analysis through several protein databases to identify evolutionary relationships, enriched molecular functions and biological processes. Flow cytometry and immunohistochemistry of tumor specimens were performed to confirm expression of druggable cell-surface antigens (Figure 1).
RESULTS: We have observed higher proliferation rates in 2D and 3D cultures and greater resistance to anti-tumor immune response in fusion-positive RMS compared to fusion-negative RMS. Surfaceome profiling of RMS cells revealed unique protein signatures for each fusion status. We have identified more than 1,500 cell-surface proteins in RMS and more than 40% of proteins are validated to be at the plasma membrane as the primary subcellular compartment. Using peptide spectral counts, we uncovered a cell surface signature 10-fold more abundant in FP-RMS than in FN-RMS. Functional classification revealed enrichment of proteins involved in immune regulation as well as metabolic pathways. Interestingly, protein expression profiles of FP-RMS vs FN-RMS are dominated by neurogenesis-associated proteins. Overexpression of top proteins identified by proteomics in FP-RMS and FN-RMS versus normal muscle has been confirmed by immunohistochemistry of patient-derived tumor tissues.
CONCLUSIONS: We have developed a unique high-throughput approach to characterize cell-surface protein signatures of RMS tumors. RMS displays a distinctive surfaceome compared to normal muscle, and fusion status drives a specific transcriptional program of cell-surface proteins. This work provides novel insights in regards to RMS development and progression. Top cell-surface candidates represent promising druggable targets, and development of novel therapeutic approaches will expand the therapeutic landscape of children diagnosed with RMS. 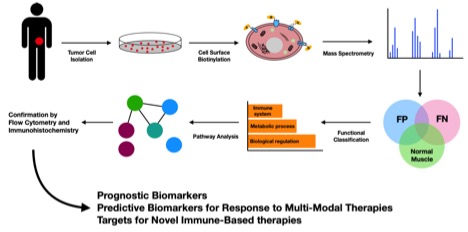
Back to 2019 Abstracts




