Genetic Analysis of a 78-patient PUV Cohort
Neeta D'Souza, BA1, Austin Thompson, BA2, Louisa Pyle, MD, PhD3, Jason Van Batavia, MD4, Dana A. Weiss, MD4, Frank Mentch, PhD4, Joseph T. Glessler, PhD4, Hakon Hakonarson, MD, PhD5, John K. Weaver, MD6.
1SUNY Downstate College of Medicine, Brooklyn, NY, USA, 2Case Western School of Medicine, Cleveland, OH, USA, 3Children's National, Washington D.C., DC, USA, 4Children's Hospital of Philadelphia, Philadelphia, PA, USA, 5Children's Hospital of Philadelphia, Philadelphia, NY, USA, 6UH Rainbow Babies and Children's Hospital, Cleveland, PA, USA.
BACKGROUND: It has long been postulated that there is a hereditary component to the development of posterior urethral valves (PUV), but the specific genetic etiology has not been identified. Prior studies have identified several copy number variants of genes associated with mesonephric duct development as well as cell-cell communication and signaling in embryogenesis. Additionally, PUV patients with copy number variants have been shown to have worse outcomes (increased rates and acceleration of renal failure) than those without any associated genetic anomalies. We present, to our knowledge, the largest PUV cohort to be evaluated for CNVs. We hypothesized that our analysis would provide an opportunity for gene discovery with the implication of understanding the genetic mechanisms responsible for PUVs.
Methods: Our study population consists of a single institution cohort of PUV patients of European origin. All patients had DNA bio-banked following IRB approval and parental consent. Controls were identified from our institutionís Center for Applied Genomics, by electronic health record mining for PUV-negative individuals. Related individuals and duplicates were removed using identity-by-state (PLINK). Genomic DNA was extracted from peripheral blood and genotyped with a variety of arrays. CNVs were called with PennCNV, and we used a segment-based scoring approach that scans the genome for CNV region containing consecutive probes with more frequent copy number changes in cases compared with controls. All the CNV-regions with nominally significant P-value<0.05 were further filtered based on quality control red flags (ParseCNV).
Results: Analysis of our cohort revealed 16 CNVs of statistical significance. The CNVs we identified have not been previously reported in the literature. See Table 1 for a full list of CNV deletions (Table 1A) and duplication (Table 1B). Two CNVs of particular interest were identified with potential links to embryologic development. The first is a deletion at chr4:108069229-108072135, a region that encodes DKK2, a protein involved in the WNT signaling pathway. The second is a deletion at chr2:203295790-203295790, a region that encodes BMPR2, a TGF-beta superfamily receptor for BMP ligands involved in kidney, bladder, and prostate development.
CONCLUSIONS:We present a genetic analysis of a 78 patient PUV cohort, identifying 16 novel CNVs that may be pathologically associated with PUV. We identified 2 CNVs containing genes involved in canonical embryologic signaling pathways. These genes will serve as the foundation for further analyses including genome-wide association, whole-genome, comparison to familial controls and collaboration for meta-analysis with other cohorts. 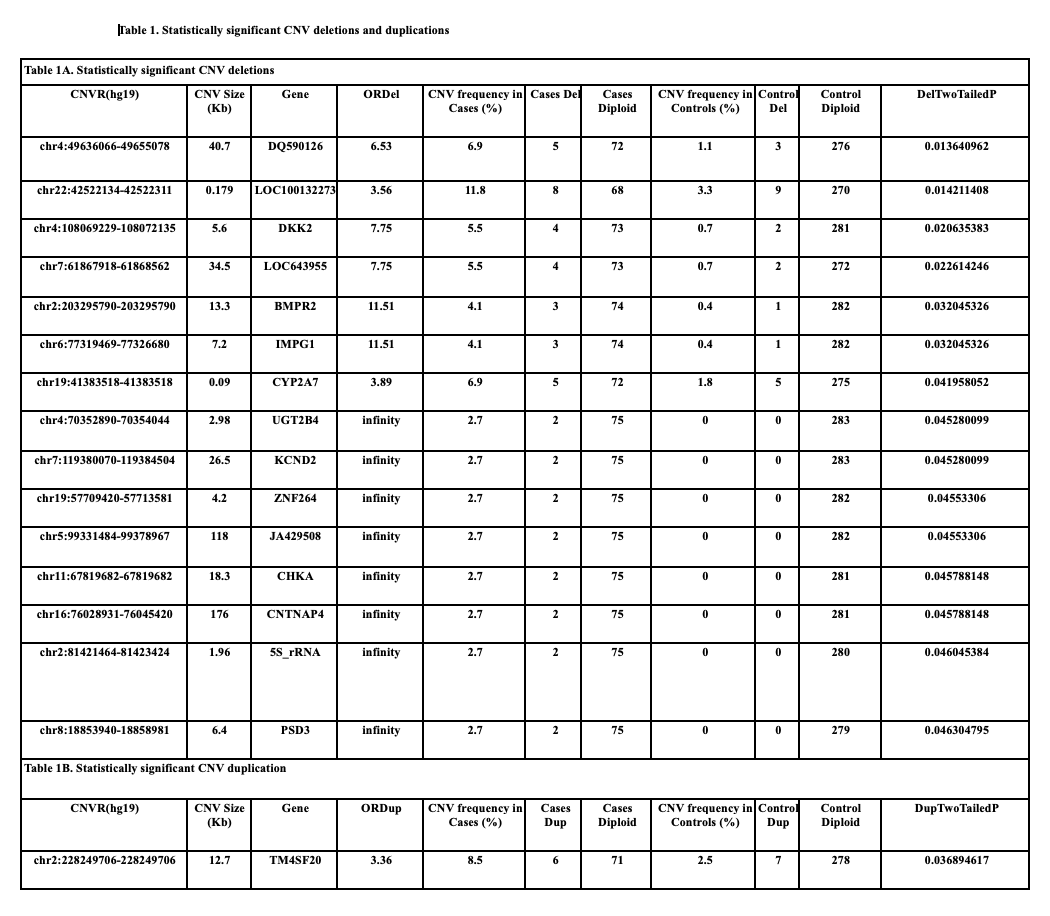
Back to 2023 Abstracts
